Abstract
Genomic and gene expression studies of multiple myeloma (MM) revealed frequent presence of common oncogenic mutations characterized in other cancers. Many mutations have corresponding targeted agents that are approved for cancers bearing them. The Multiple Myeloma Research Foundation (MMRF) and its partners launched the MyDRUG clinical trial (NCT03732703) to gain a deeper understanding of the safety and efficacy of targeting these specific pathways in MM. MyDRUG is a genomically-guided umbrella trial for relapsed/refractory myeloma patients (RRMM) who received at least one, but no more than three, prior therapies, have been exposed to both a PI and an IMiD, and had early relapse after initial treatment. Patients enrolling in MyDRUG are genomically profiled for 'actionable’ tumor abnormalities: activating or oncogenic mutations in CDKN2C, FGFR3, KRAS, NRAS, BRAF V600E, IDH2 or the t(11;14) translocation. Patients with >30% mutated allele frequency in any of the listed genes can be enrolled to one of the treatment arms with therapeutic agents directed against the abnormal gene or associated activated pathway. Patients that lack such a defined abnormality or have allelic burdens below the 30% threshold are assigned to a "non-actionable” treatment arm.
In this study, patients with activating mutations in NRAS, KRAS or BRAF ("MAPK Pathway") were assigned to sub-protocol C1 and treatment with Cobimetinib, a MEK1/2 inhibitor approved for the treatment of melanoma. Cobimetinib plus Dexamethasone were administered for two 28-day cycles followed by the addition of an Ixazomib, Pomalidomide and Dexamethasone (IPd) backbone therapy for all subsequent treatment cycles until disease progression. Reports from this and other studies of MAPK pathway inhibitors in myeloma indicate there is clinical activity. Bone marrow samples were collected from patients prior to therapy with Cobimetinib (baseline/BL), after 2 cycles of Cobimetinib therapy (EOC2), after 2 cycles of combination with IPd (EOC4) and at the end of treatment or at disease progression (EOT). All samples were processed into CD138+ myeloma cell and CD138- "immune cell” fractions for single-cell analysis.
In this study, the CD138- "immune cell” fractions were transcriptionally profiled by single-cell RNA-sequencing (scRNA) at up to four time points (Figure 1A). The CD138- fraction also contained residual CD138+ myeloma blasts, enabling detailed single-cell analysis of the changes in both the immune and tumor cell compartments of the bone marrow from RRMM patients over time and therapy.
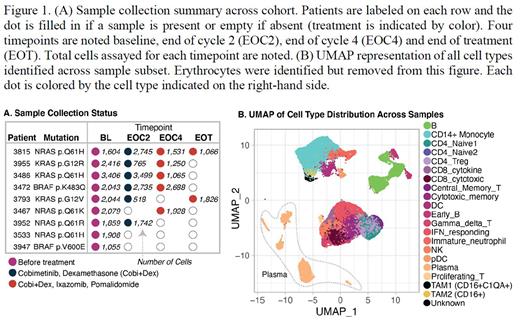
Transcriptome profiles (~50,000 cells) were generated across 22 samples from 9 patients with tumor MAPK Pathway mutations at entry: BRAF (p.K483Q, p.V600E, n=2); KRAS (p.Q61H, p.G12V, p.G12R, n=3); NRAS (p.Q61H, p.Q61R, p.Q61K, n=4). Immune cell types assayed include NK and T cells, CD14+ and CD16+ Monocytes, B cells, Neutrophils, and Dendritic cells, listed in order of sampling size (Figure 1B) with an average of 1,691 immune cells sampled per patient timepoint (excluding erythrocytes). Over 4,000 plasma cells were captured with an average of 200 plasma cells per patient timepoint.
Several changes in both the immune and plasma cell compartments were observed across timepoints. Within the monocyte/macrophage lineage, tumor associated macrophage (CD16+ TAM) subsets increased over the course of treatment from BL to EOC4. Further, TAM populations exhibit differences in expression of the major histocompatibility complex class II suggesting differential regulation of pro-inflammatory signaling over the course of treatment. Gene expression changes were observed in both the immune and plasma cell compartments between BL and EOC2: Increased IFN-signaling was observed in the T-cell compartment suggesting induction of T-cell activity in response to the MEK-inhibitor regimen. In the residual plasma cells compartment, two plasma cell clusters differed in expression of a transcriptional signature associated with MAPK pathway activation (MAPK). After exposure to Cobimetinib, the plasma cell clusters that expressed higher levels of this MAPK signature decreased in proportion relative to clusters with lower expression of MAPK. In summary, here we report on our initial observations of dynamic transcriptional changes in specific immune and myeloma cell compartments that are associated with targeting the MAPK pathway in the MyDrug C1 sub-protocol.
Disclosures
Jayasinghe:WUGEN: Consultancy; MMRF: Consultancy. Gnjatic:Bristol Myers Squibb: Research Funding; Genentech: Research Funding; Boehringer-Ingelheim: Research Funding; Celgene: Research Funding; Janssen R&D: Research Funding; Pfizer: Research Funding; Regeneron: Research Funding. Cho:BMS/Celgene: Other: Receive laboratory research support from the above companies. Salary value is less than $10,000 per company., Research Funding; Takeda: Other: Receive laboratory research support from the above companies. Salary value is less than $10,000 per company., Research Funding. Kumar:AbbVie,: Membership on an entity's Board of Directors or advisory committees, Research Funding; Celgene: Membership on an entity's Board of Directors or advisory committees, Research Funding; Janssen,: Membership on an entity's Board of Directors or advisory committees, Research Funding; Takeda,: Membership on an entity's Board of Directors or advisory committees, Research Funding; Adaptive,: Membership on an entity's Board of Directors or advisory committees, Research Funding; KITE,: Research Funding; MedImmune/Astra Zeneca,: Membership on an entity's Board of Directors or advisory committees, Research Funding; Merck,: Research Funding; Novartis,: Research Funding; Roche: Research Funding; Sanofi: Research Funding; Oncopeptides: Other: Independent review committee.
OffLabel Disclosure:
Cobimetinib is a reversible inhibitor of mitogen-activated protein kinase (MAPK)/extracellular signal regulated kinase 1 (MEK1) and MEK2. MEK proteins are upstream regulators of the extracellular signal-related kinase (ERK) pathway, which promotes cellular proliferation. BRAF V600E and K mutations result in constitutive activation of the BRAF pathway which includes MEK1 and MEK2.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal